Welcome to the CRISPR Vision Program! Funded by the National Institutes of Health (NIH), we're innovating non-viral gene editing therapies for inherited retinal disorders. Our work aims to bring the groundbreaking potential of CRISPR technology to those affected by these diseases.
About Our Program
We're pushing the frontiers of gene editing technology. Our focus lies in overcoming the limitations of existing viral delivery systems and introducing novel, non-viral treatments for inherited eye diseases, particularly those affecting the retinal pigment epithelium.
Our Innovative Approach
Our novel non-viral delivery systems — synthetic nanoparticles and Target Active Gene Editors (TAGE) — sit at the heart of our mission. These promising technologies aim to target the retina efficiently and treat rare conditions like Best Disease and Leber Congenital Amaurosis.
The Impact
We're pushing the boundaries of gene editing by developing treatments for inherited retinal disorders. Our work could pave the way for treating numerous inherited retinal disorders, marking a significant stride in the fight against genetic eye diseases.

About the Somatic Cell Genome Editing (SCGE) Consortium
As a part of the SCGE Consortium, we are working in a community dedicated to developing safe and effective gene editing methods to treat genetic diseases in somatic cells. Click here for more information about the SCGE NIH Working Group and the Common Fund.
Team
Over 30 members within the program. The leadership team consists of:
Press
"Innovations in gene-editing treatment in Wisconsin and abroad," The Morning Show, Wisconsin Public Radio, November 28, 2023, Link Listen
"NIH awards more than $140 million to help accelerate genome editing approaches from the lab to the clinic," National Institutes for Health, Link
"UW researchers will develop gene editing therapy to treat blindness," University of Wisconsin-Madison, Link
Associated articles:
Wisconsin State Journal, Link
National Eye Institute Director, Link
Foundation for Fighting Blindness, Link
"New gene-editing technique holds potential for treating childhood blindness," University of Wisconsin-Madison, Link PDF
"Rising Sparks: Marcela Tabima, Regenerative Biology" University of Wisconsin-Madison, Link
"These 10 scientists are leading a new generation of gene editors developing CRISPR medicines" STAT, Link PDF
Selected Publications
Gowen BG, Khekare P, McCawley SR, Melton K, Soares C, Chan J, Jani V, Boivin P, Bans A, Leong WI, Cantor AJ, Walleshauser J, Otoupal PB, Mepani RJ, Silverman AP, Haak-Frendscho M, Wei SC. Identification of a Guide RNA Targeting an Ultraconserved Element for Evaluation of Cas9 Genome Editors Across Mammalian Species. CRISPR J. 2024 Sep 23. doi: 10.1089/crispr.2024.0053. Epub ahead of print. PMID: 39308435. Link PDF
Kabra, M., Shahi, P. K., Wang, Y., Sinha, D., Spillane, A., Newby, G. A., Saxena, S., Tong, Y., Chang, Y., Abdeen, A. A., Edwards, K. L., Theisen, C. O., Liu, D. R., Gamm, D. M., Gong, S., Saha, K., & Pattnaik, B. R. (2023). Nonviral base editing of KCNJ13 mutation preserves vision in a model of inherited retinal channelopathy. The Journal of Clinical Investigation. Link Press
Sinha, D., Steyer, B., Shahi, P. K., Mueller, K. P., Valiauga, R., Edwards, K. L., Bacig, C., Steltzer, S. S., Srinivasan, S., Abdeen, A., Cory, E., Periyasamy, V., Siahpirani, A. F., Stone, E. M., Tucker, B. A., Roy, S., Pattnaik, B. R., Saha, K., & Gamm, D. M. (2020). Human iPSC Modeling Reveals Mutation-Specific Responses to Gene Therapy in a Genotypically Diverse Dominant Maculopathy. The American Journal of Human Genetics, 107(2), 278–292. LinkSaha, K., Sontheimer, E. J., Brooks, P. J., Dwinell, M. R., Gersbach, C. A., Liu, D. R., Murray, S. A., Tsai, S. Q., Wilson, R. C., Anderson, D. G., Asokan, A., Banfield, J. F., Bankiewicz, K. S., Bao, G., Bulte, J. W. M., Bursac, N., Campbell, J. M., Carlson, D. F., Chaikof, E. L., … SCGE Consortium. (2021). The NIH Somatic Cell Genome Editing program. Nature, 592(7853), 195–204. Link
Chen, G., Abdeen, A. A., Wang, Y., Shahi, P. K., Robertson, S., Xie, R., Suzuki, M., Pattnaik, B. R., Saha, K., & Gong, S. (2019). A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nature Nanotechnology, 14(10), 974–980. Link
Wang, Y., Shahi, P. K., Xie, R., Zhang, H., Abdeen, A. A., Yodsanit, N., Ma, Z., Saha, K., Pattnaik, B. R., & Gong, S. (2020). A pH-responsive silica-metal-organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome-editing machineries. Journal of Controlled Release, 324, 194–203. LinkSamimi, K., Pattnaik, B. R., Capowski, E. E., Saha, K., Gamm, D. M., & Skala, M. C. (2022). In situ autofluorescence lifetime assay of a photoreceptor stimulus response in mouse retina and human retinal organoids. Biomedical Optics Express, 13(6), 3476–3492. Link
Wang, Y., Ma, B., Abdeen, A. A., Chen, G., Xie, R., Saha, K., & Gong, S. (2018). Versatile Redox-Responsive Polyplexes for the Delivery of Plasmid DNA, Messenger RNA, and CRISPR-Cas9 Genome-Editing Machinery. ACS Applied Materials & Interfaces. Link
Steyer, B., Bu, Q., Cory, E., Jiang, K., Duong, S., Sinha, D., Steltzer, S., Gamm, D., Chang, Q., & Saha, K. (2018). Scarless Genome Editing of Human Pluripotent Stem Cells via Transient Puromycin Selection. Stem Cell Reports, 10(2), 642–654. Link
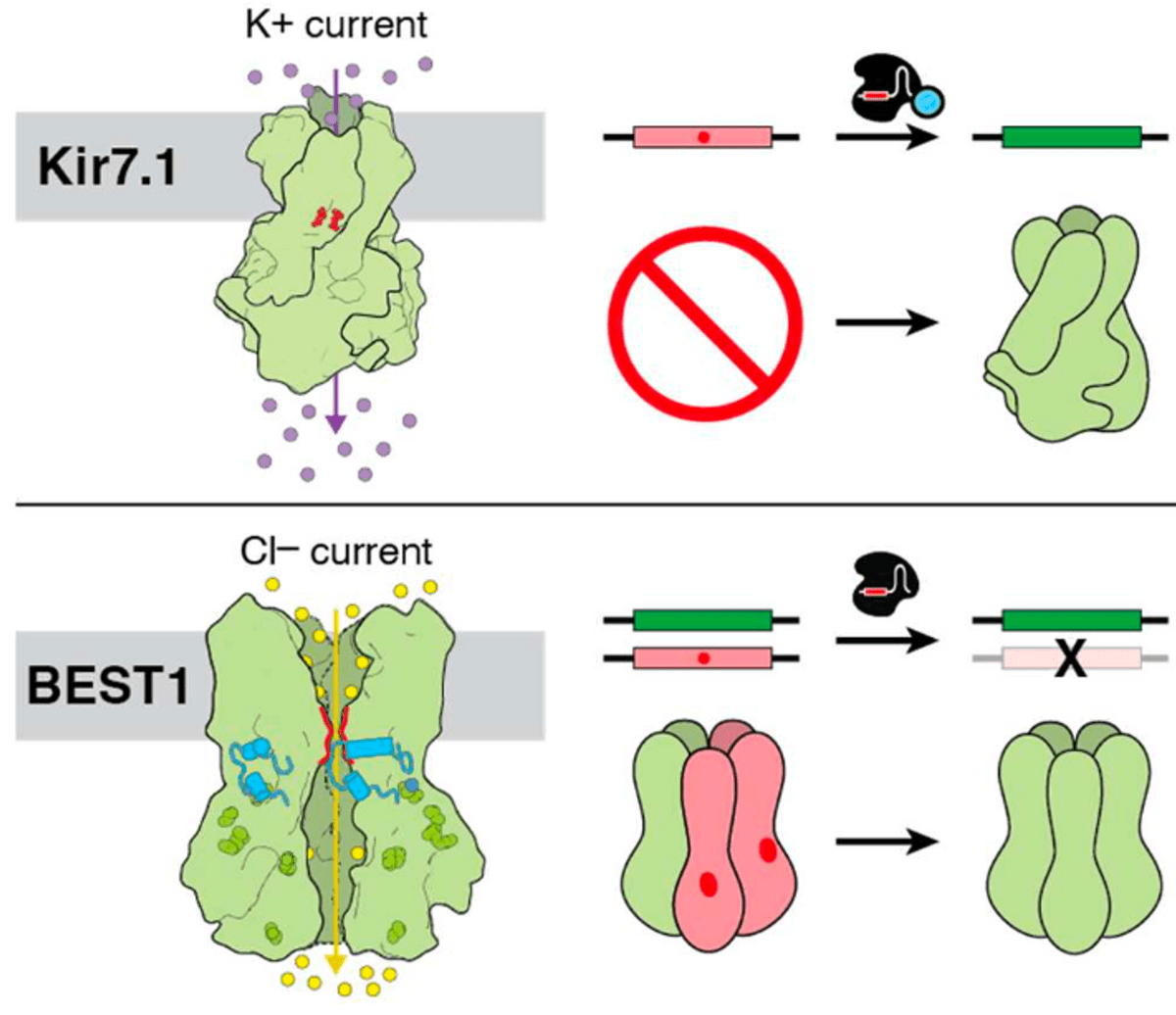
Overview of the genome editing strategy for two rare inherited diseases: Leber congenital amaurosis (top) and Best disease (bottom). Channel proteins in the cell membrane of retinal cells are shown on the left. The gene editing strategy on the right will modify the mutant gene in order to fix the mutations and establish functional channel proteins.
Additional technical information is here.

Progress and Goals
Keep up with our progress. We're committed to developing our non-viral gene editing therapies, creating a platform to tackle various rare eye diseases, and providing clarity on the development path for new gene-editing therapies. Stay tuned for updates on our journey to turn the promise of CRISPR technology into a reality.